Why Simone grows spheroids in 3D for researching chromatin domains

Simone Sidoli
www.sidolilab.org
Information:
Professor
Scientific Director of the Proteomics Core
Albert Einstein College of Medicine
New York
Reason for using clinostat
We investigate the accessibility and the dynamics of chromatin domains in health and disease. Chromatin decondensation is a shared phenomenon in aging, different types of cancer and infections. Understanding which chromatin domains are being disordered during a reduction in cell fitness could have remarkable implications in delaying or arresting this phenomenon. For instance, more specific treatments could be developed if inhibitors are developed against proteins interacting exclusively with disordered chromatin.
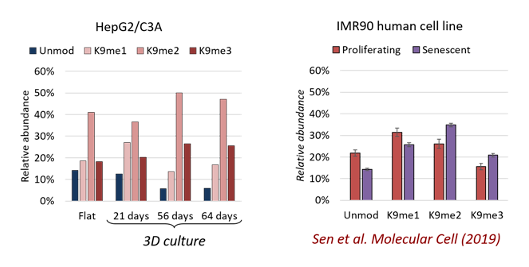
Our methods are not applicable on animals or biopsies, because chromatin accessibility and transcription is performed via metabolic labelling feasible only with cell cultures. We utilize the CelVivo system because canonical cell cultures are excessively involved in DNA replication, the cell cycle and they have minimal cell-Mature chromatin has wider domains of silenced heterochromatin. This is because most cells in a solid tissue are in a senescent stage. We can accumulate senescent markers such as K9me2/me3 on histone H3 by growing spheroids for a longer period of time on the CelVivo bioreactor without affecting their fitness.
Learnings/Tips and Tricks
We have numerous projects involving the growth of spheroids, and our user list is growing. Currently, we are amazed about how simple is to maintain and prevent contaminations of these spheroids also for inexperienced users. Please, feel free to visit our website and write to us if you work in the United States and you are interested in seeing the system in action. The Sidoli lab is currently the training site and Center of Excellence of CelVivo in the United States.
“We have numerous projects involving the growth of spheroids, and our user list is growing”.
WEBINAR
Chromatin Homeostasis
In this presentation, Professor Simone Sidoli from the Albert Einstein College of Medicine discusses his research about how he has been modeling accessible heterochromatin to identify proteins and histone modifications regulating chromatin homeostasis.
Highlighted Research
Get inspired by Simone and his group’s research made with the ClinoStar system.
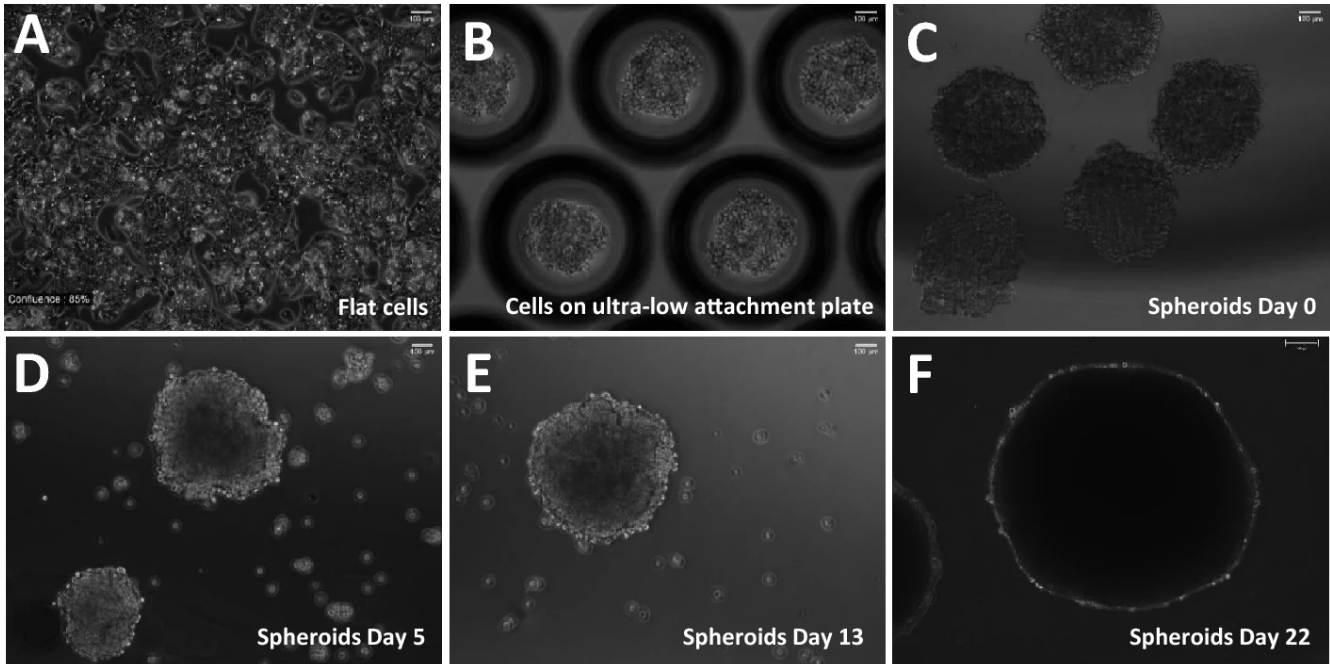
Global Level Quantification of Histone Post-Translational Modifications in a 3D Cell Culture Model of Hepatic Tissue
DOI: 10.3791/63606
Presented below is a workflow for modeling, treating, and analyzing quiescent chromatin modifications using a three-dimensional (3D) cell culture system.
Investigation of reversible histone acetylation and dynamics in gene expression regulation using 3D liver spheroid model
DOI: 10.1186/s13072-022-00470-7
A demonstration, that 3D liver spheroids are a suitable system to model chromatin dynamics and response to epigenetics inhibitors.
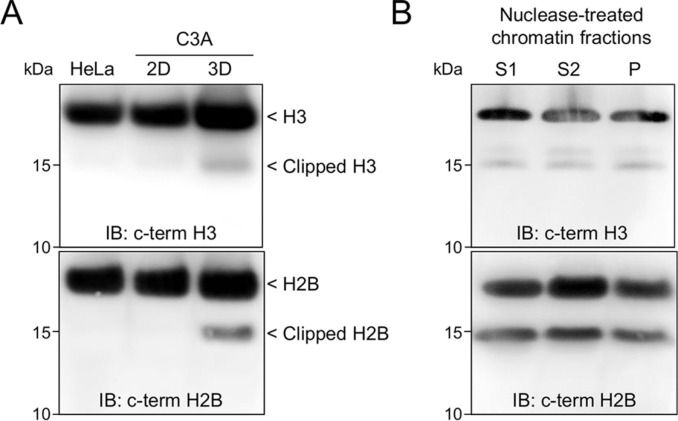
Top-down and Middle-down Protein Analysis Reveals that Intact and Clipped Human Histones Differ in Post-translational Modification Patterns
DOI: 10.1002/btpr.3253
Using top-down and middle-down protein analysis by mass spectrometry, we report histone H2B and H3 N-terminal tail clipping in human hepatocytes and demonstrate a relationship between clipping and co-existing PTMs of histone H3.
Read more ClinoStar publications
Start optimising your
In Vitro models today

